- Home
- GEP
- Genomes
- Genome Browser
- Tools
- My Data
- Help
Track Hub Filters Quick Start Guide
Track Hubs are a method of displaying remotely-hosted annotation data quickly and flexibly on any
UCSC assembly or remotely-hosted sequence. There are different filtering options available for
bigBed files depending on the kind of data the filter will be applied to. These filters are also
described in the trackDb help doc.
Note: for configurable features, like filters, an additional period "." or plus
"+" is required in the type declaration, for instance type bigBed 5 .
or type bigBed 9 +.
Contents
filter.fieldName - Used for numerical data
filterText.fieldName - Used for text filtering
filterValues.fieldName - Used for filtering by prespecified values or categories in data
filter.fieldName
filter.fieldName is used to enable numerical filtering within a
field or column. It is often seen as a filter on data that contains a score field.
It requires a default parameter to be passed, often this parameter is 0.
By default, the range of values will be 0 to 1000. However, this range can be modified with the
filterLimits.fieldName parameter. Additionally, the filter can be modified to take in a
range of values with the filterByRange.fieldName on parameter.
For more information on filter.fieldName, see the trackDb help doc entry.
filter Example 1
In this first example, we have a simple track with 10 items. The data looks as follows:
chr7 127000000 127000005 1 1 chr7 127000010 127000015 2 2 chr7 127000020 127000025 3 3 chr7 127000030 127000035 4 4 chr7 127000040 127000045 5 5 chr7 127000050 127000055 6 6 chr7 127000060 127000065 7 7 chr7 127000070 127000075 8 8 chr7 127000080 127000085 9 9 chr7 127000090 127000095 10 10
In this case, we have duplicated the name and score fields for
clarity. We will be applying a default filter of 4 to the score field. This
will mean that by default only items 4-10 will display. This filter is enabled with the
line filter.score 4. The hub.txt looks as follows:
track filterScore4 shortLabel filter.fieldNameDefault 4 longLabel Numerical filter with a default value of 4 passed visibility pack type bigBed 5 . filter.score 4 bigDataUrl example1.bb
Below are all the materials for example 1:
The example session will display an image like the following, which hides items 1-3 and
displays items 4-10. The three filtered items are also noted on the longLabel above the
track as (3 items filtered). If we just want to enable filtering, but pass no
default value, we can use filter.score 0.

filter Example 2
In this second example we have four tracks with filter.fieldName to
allow numerical filtering, filterByRange.fieldName on
to enable range filtering, and filterLimits.fieldName to designate upper
and lower range boundaries. filter.fieldName will be used on two separate fields
(score and name) to demonstrate multiple filters. Lastly, filterLabel.fieldName
will be used to change the default filter message to instead "Value range to filter"
for the score field. The data used is the same as example 1 above:
chr7 127000000 127000005 1 1 chr7 127000010 127000015 2 2 chr7 127000020 127000025 3 3 chr7 127000030 127000035 4 4 chr7 127000040 127000045 5 5 chr7 127000050 127000055 6 6 chr7 127000060 127000065 7 7 chr7 127000070 127000075 8 8 chr7 127000080 127000085 9 9 chr7 127000090 127000095 10 10
The data are organized as a bed5. Standard filtering will be enabled in the 4th name
field, and filtering by ranges on the 5th score field. The score field
will also be given a custom label with the filterLabel parameter. Here is an
example of the first track stanza:
track filteringByRangeAllValues shortLabel filteringByRangeDefault longLabel Filter by range enabled with default score including all values visibility pack type bigBed 5 . filter.name 0 filter.score 0:10 filterByRange.score on filterLimits.score 0:10 filterLabel.score Value range to filter bigDataUrl example2.bb
Below are all the materials for example 2:
Enabling range filters for the score field as well as the standard filter for
the name field, we see the following options in the track description page.
Any number of filters can be enabled on a track simultaneously.

Going to the example session will display a browser image like so:
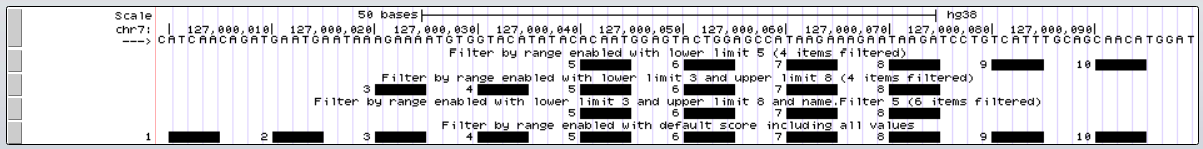
Each of the tracks is filtering by a different value, and in the final track two separate filters are enabled.
- The first track (5-10) displays only items with scores 5-10.
- The second track (3-8) displays only items with scores 3-8.
- The third track has two filters. A range filter (3-8) and a second filter (5). Only the values that pass both filters are seen.
- The fourth track has a filter inclusive of all values passed(0-10), so all items are displayed.
filter Example 3
This third example explores how the numerical filters interact with non-numerical characters.
The data is comprised of 10 items as a bed5, with the same coordinates as the examples above, a name field, and an arbitrary score field. The name field contains a mix of numerical and non-numerical characters.
chr7 127000000 127000005 0 0 chr7 127000010 127000015 -1 0 chr7 127000020 127000025 2% 0 chr7 127000030 127000035 -3 0 chr7 127000040 127000045 4 0 chr7 127000050 127000055 5n 0 chr7 127000060 127000065 5 0 chr7 127000070 127000075 NA 0 chr7 127000080 127000085 . 0 chr7 127000090 127000095 <> 0
Filtering will be enabled on the 4th name field.
The trackDb stanza looks as follows:
track filteringNonNumerical shortLabel filteringNonNumerical longLabel Using numerical filters on a field with both numerical and non-numerical values visibility pack type bigBed 5 . filter.name 0 bigDataUrl example3.bb
Below are all the materials for example 2:
The filter is being passed on the name field, with a default value of 0. The
example session will show which items still display with this default value:

The only items being filtered are the negative values, -1 and -3. Entirely non-numerical characters are interpreted as 0. If we instead change the filter to be 2, we see the following:

In this case we see the items that start with non-numerical characters get filtered. Items
that start with a number, and are following by another character, are treated as the number.
This can be seen with the 2% value, which remains visible with the filter active.
It is important to keep in mind that non-numerical values, such as NA, will be visible
when the default 0 filter is active, but will be removed when any positive numerical filter
is activated.
filterText.fieldName
filterText.fieldName is used to enable text searching in the specified fieldName.
This will display any items passed which match exactly the searched term, or only part
of the search term. Two types of searching are supported, wildcard searching (*) or regular
expression searching (regexp). The mode between the two types can be freely changed in the track
description page, or a default passed using the filterType.fieldName parameter. Lastly,
the filter label will be the description of the field as specified by the autoSql (.as) file.
This label can be customized with the filterLabel.fieldName parameter.
A value can be passed with this setting to enable a specific filter by default. Also, the default search type is wildcard (*).
For more information on filterText.fieldName, see the trackDb help doc entry.
filterText Example 1
In this first example, we have 10 genes with arbitrary coordinates as follows:
chr7 127000000 127000005 EGFR 1 chr7 127000010 127000015 VEGFA 2 chr7 127000020 127000025 APOE 3 chr7 127000030 127000035 IL6 4 chr7 127000040 127000045 TGFBI 5 chr7 127000050 127000055 BRCA1 6 chr7 127000060 127000065 BRCA2 7 chr7 127000070 127000075 MTHFR 8 chr7 127000080 127000085 ESR1 9 chr7 127000090 127000095 AKT1 10
By default, we would like our data to display only BRCA1 and BRCA2. The easiest way to accomplish
this is to enable a filterText wildcard filter. We will be filtering on the
name field using the following setting: filterText.name BRCA*
The hub.txt looks as follows:
track filterTextDefaultBRCA shortLabel filterTextBRCA longLabel Wildcard filterText with default BRCA value visibility pack type bigBed 5 . filterText.name BRCA* bigDataUrl filterTextExample1.bb
Below are all the materials for example 1:
The example session will display an image like the following, showing only the BRCA items.
The eight filtered items are also noted on the longLabel above the
track as (8 items filtered). If we just want to enable filtering, but pass no
default value, we can use filterText.name *.

We can also change the filter type or filter value (or remove values) by going to the track description page, which will show the following:
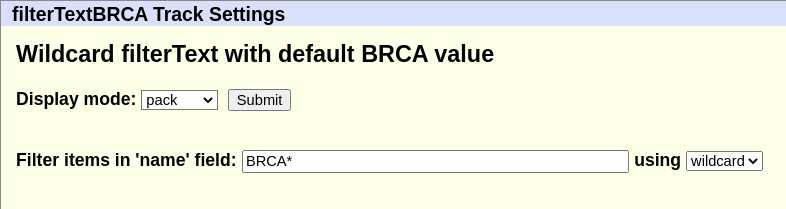
Changing the filter from BRCA* to *A* would expand the wildcard
match to all items with A.
See the trackDb help doc entry for
additional information including an example using regexp. For instance, with wildcard changed to a
regexp type of search, putting in .*A\|B.* will match any items with an A or B in it,
while .*[0-9] will match any item ending in a number.
filterValues.fieldName
filterValues.fieldName is used to enable filtering by pre-specified values within
a field. It can be used on fields that can contain one text value or a list of comma-separated
values of text, like "classA,classB". Usually, these are category names. The option requires
at least one value to filter on.
Every individual possible value that can ever occur in the field must be passed in a comma
separated list. You will then be able to select those values as categories, choosing to display
only items that belong to one, any, or at least one of the selected values. By default, the user
can select multiple values from this list and the filter lets pass any features with at least
one of these values (multipleListOr). The type of selection can be designated by
passing the optional filterType.fieldName parameter. Possible options are:
single- Allows selection of a single item from dropdown menu.singleList- Allows selection of a single item from dropdown menu. Accepts comma-separated values in the bigBed field.multiple- Allows selection of any number of items from dropdown menu. Shows items that contain at least one of the selections.multipleListOr- Allows selection of any number of items from dropdown menu. Shows items that contain at least one of the selections. Accepts comma-separated values in the bigBed field. Enables the radio button that allows swapping of filter type in the track description page.multipleListOnlyOr- Same asmultipleListOrexcept it disables the option to change the filter type from the browser interface.multipleListAnd- Allows selection of any number of items from dropdown menu. Displays only items containing all of the selections. Accepts comma-separated values in the bigBed field. Enables the radio button that allows swapping of filter type in the track description page.multipleListOnlyAnd- Same asmultipleListAndexcept it disables the option to change the filter type from the browser interface.
As with other filters, default values can be passed to filterValues using the
filterValuesDefault.fieldName parameter. It can take a comma-separated list
just like filterValues.fieldName, and any items included will be automatically selected.
The labels in the menu shown to the user can be configured to display a different name/label than the one present in the bigBed field. This can be helpful when the data values are written in short form, but you want a longer more descriptive name to show up in the UI. The format for this substitution is as follows:
filterValues.fieldName fieldValue1|alternativeName1,fieldValue2|alternativeName2...
In this example, a bigBed could have value AML in a field called Disease, but we would
like the menu to display Acute Myeloid Leukemia so the line could be
filterValues.Disease AML|Acute Myeloid Leukemia,MSC|Melanoma Skin Cancer... where
the filter display would have the full disease names, while the data instead in the bigBed
was the abbreviation. This can also be used to reduce the size of bigBed files.
The
following session contains a hub with example tracks of all
the possible filterValues settings. It can be used to explore the differences
and restrictions of each of the settings.
filterValues Example 1
In this first example, we have a small track with 10 items. The score and names of the items are the same, and there is an additional field added which labels the score as either an even or odd number, making our file a bigBed 5+1:
chr7 127000000 127000005 1 1 odd chr7 127000010 127000015 2 2 even chr7 127000020 127000025 3 3 odd chr7 127000030 127000035 4 4 even chr7 127000040 127000045 5 5 odd chr7 127000050 127000055 6 6 even chr7 127000060 127000065 7 7 odd chr7 127000070 127000075 8 8 even chr7 127000080 127000085 9 9 odd chr7 127000090 127000095 10 10 evenWe wish to add a filter which allows selection on whether the item is even or odd. This is enabled by the line
filterValues.OddEven odd,even. Remember that all
possible values need to be listed. We also wish for only a single selection
to be possible, which we will do with the following setting
filterType.OddEven singleList. Below is the track stanza in the hub.txt:
track filteValuesOddEven shortLabel filterValues.OddEven odd,even longLabel filterValues categorical filter on odd and even values visibility pack type bigBed 5 + 1 filterValues.OddEven odd,even filterType.OddEven singleList bigDataUrl filterValuesExample1.bb
Below are all the materials for this example:
filterValues Example 2
In this second example, we have signal data on 10 items, and want to enable filtering on any number of annotation type. Note that the annotation type is written in shorthand:
chr7 127000000 127000005 signal1 0 DNA-BR,AH chr7 127000010 127000015 signal2 0 AS,BS chr7 127000020 127000025 signal3 0 BS chr7 127000030 127000035 signal4 0 BS chr7 127000040 127000045 signal5 0 DNA-BR chr7 127000050 127000055 signal6 0 DNA-BR,BS chr7 127000060 127000065 signal7 0 AS chr7 127000070 127000075 signal8 0 BS chr7 127000080 127000085 signal9 0 AS,AH chr7 127000090 127000095 signal10 0 DNA-BR,BS
In this case, the annotations (the sixth annotationType column) represent the following:
- DNA-BR - DNA-binding region
- AS - active site
- AH - alpha helix
- BS - beta strand
Some signals contain more than one type of annotation. We will enable the categorical filter
using the filterValues.annotationType setting, including all the possible values.
We will also substitute the complete annotation type names by using the pipe "|" character.
We work with researchers most interested in DNA-binding regions that are also beta strands,
so we will want the default behavior to display only those items. For this we will use
filterType.annotationType multipleListAnd to require matching on all selections,
and filterValuesDefault.annotationType DNA-BR,BS to pass the default values.
Note that we do not use the type multipleListOnlyAnd, as that would not allow
users to change selection type. Below is the complete track stanza of the hub.txt file:
track filteValuesAnnotationType shortLabel filterValues.annotationType longLabel filterValues categorical filter on multiple annotation types visibility pack type bigBed 5 + 1 filterValues.annotationType DNA-BR|DNA-binding region,AS|active site,AH|alpha helix,BS|beta strand filterType.annotationType multipleListAnd filterValuesDefault.annotationType DNA-BR,BS bigDataUrl filterValuesExample2.bb
Below are all the materials for this example:
Going to the example session will result in a browser image as such:

Note that only signal6 and signal10 are displayed by default. That is because they are the only two items that have both DNA-BR (DNA-binding region) and BS (beta strand) as their type of annotation. If we click into the track, we see additional filter options. In the image below we have switched the filter to one or more match, meaning that if the items contain any of the selected items they will display. We have also expanded the selection to include AH (alpha-helix). Lastly, note that the menu displays the alternative long names instead of the shorthand:
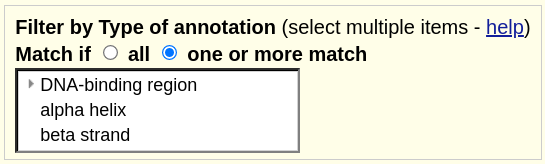
This will result in all items being displayed except for signal 7. This is because it is the only item that contains none of the selected categories, with its only annotation being AS (active site):

Additional Resources
- Track Hub User Guide
- Guide To useOneFile setting
- Search file .ix documentation
- Mailing list question with searchable Track Hub
- Mailing list question with searchable Custom Tracks
- Track Database (trackDb) searchTrix Definition
- Quick Start Guide to Organizing Track Hubs into Groupings
- Quick Start Guide to Assembly Track Hubs